Optimizing Workflows in Distributed Systems
A Case Study for the Lleida Population Cancer Registry
University of Lleida
University of Lleida
University of Lleida
Whoami
- Industrial PhD Student at GFT
- Working on industrial implementations of federated learning

Introduction
Context
This work was part of an Industrial PhD, Florensa Cazorla (2023), collaboration between the Population Cancer Registry, Arnau de Vilanova Hospital, and the University of Lleida.
Purpose
Develop an optimized platform that enables high-quality data for identifying associations between medications and cancer types within the Lleida population registry.
Team
- Dídac Florensa: PhD student, responsible for data management, analysis, and requirements gathering.
- Pablo Fraile: PhD student, responsible for implementing and developing the platform.
- Jordi Mateo: Professor at the University of Lleida, leading the project.
Problem Statement
Goal
Analyze associations: Medication and cancer type effects on patient survival (protective or harmful).
Challenge
Analyzing 79,931 combinations of medications and cancer types from 2007-2019.
Inital Approach
A single machine would require 61 days to complete this analysis, with each combination consuming 66 seconds.
Profiling
Goal
- Identify bottlenecks
- Identify inefficiencies
- Propose optimizations
Findings
- Data not retrieved in a single query
- Join-like applied on a non-relational DB
- Queries misaligned with schema structure
Proposals
- Schema redesign based on query access patterns.
Yearly Schema
ATC Code Schema
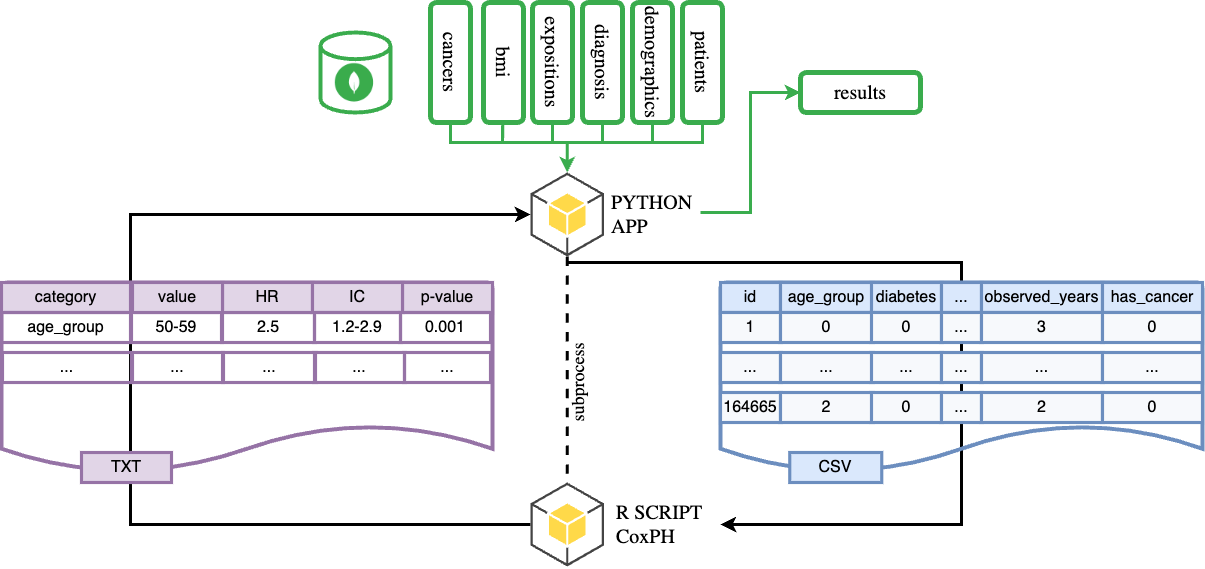
Data schema impact
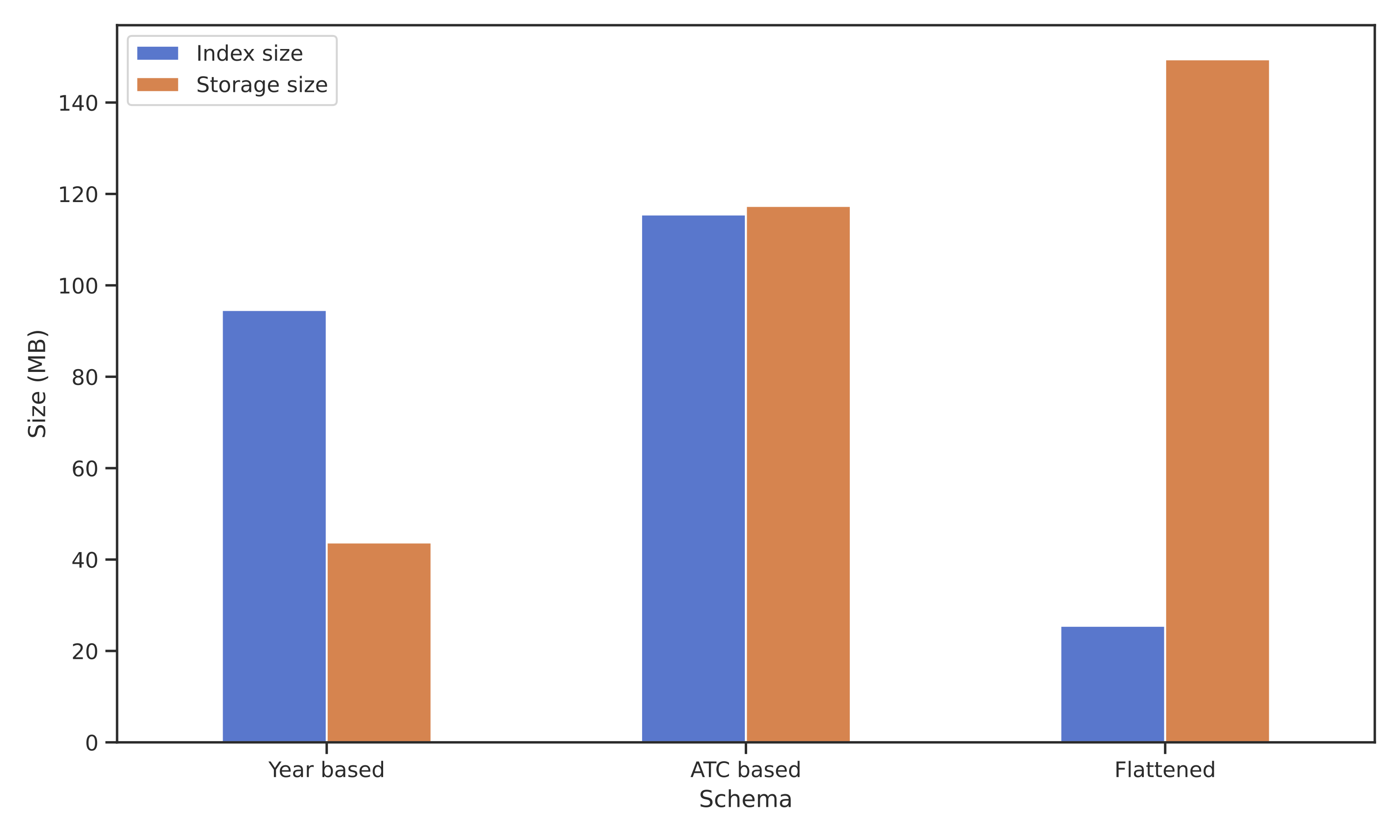
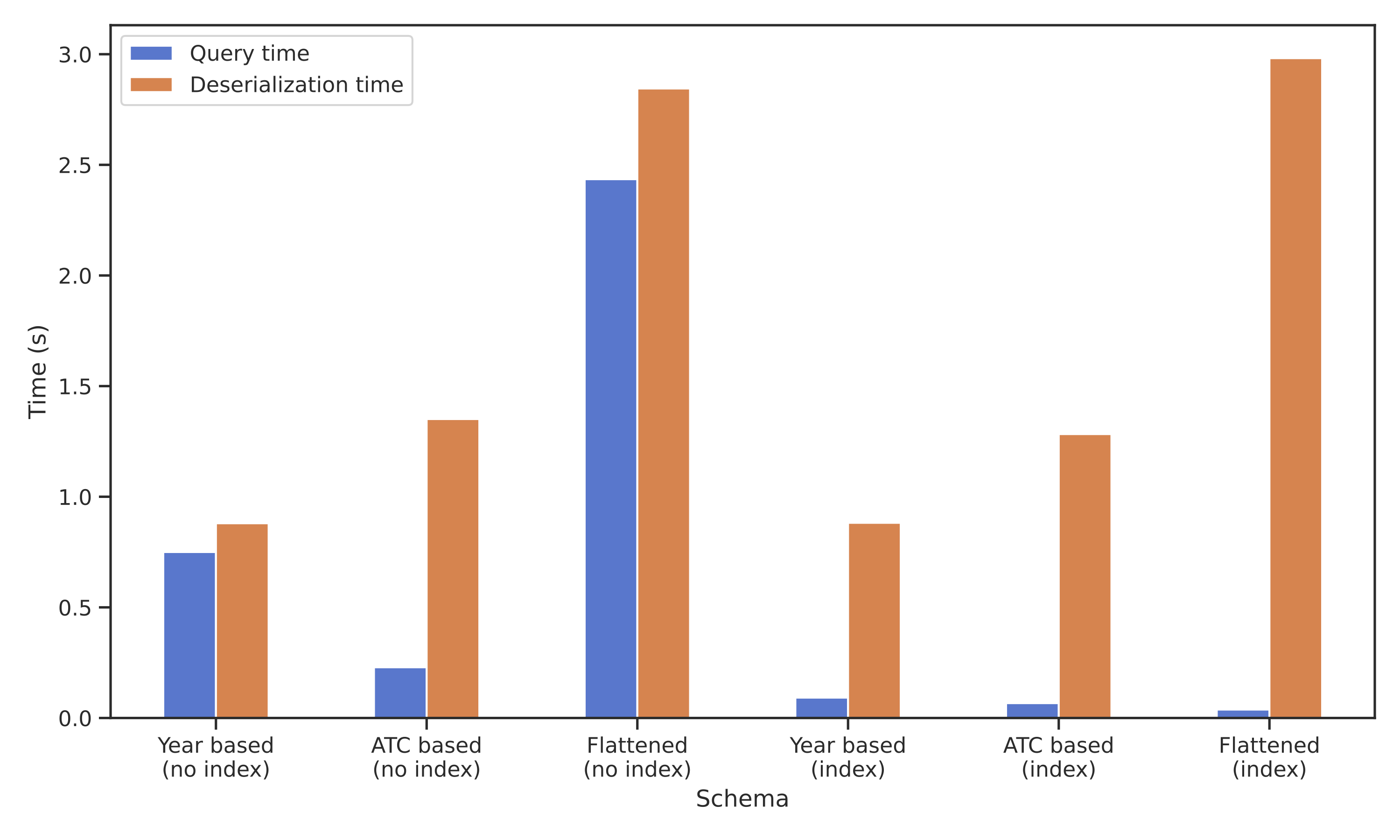
Observation
Proposed solutions reduce query time (at the cost of disk space for indexes). However, deserialization time increases across all proposals.
Next Steps
How can we simultaneously minimize deserialization time and reduce query execution?
Deserialization
Findings
- PyMongo returns Python dictionaries → slow for large result sets
- PyMongoArrow improves typing, but still memory-heavy
- Optimal performance requires columnar layout with primitive types
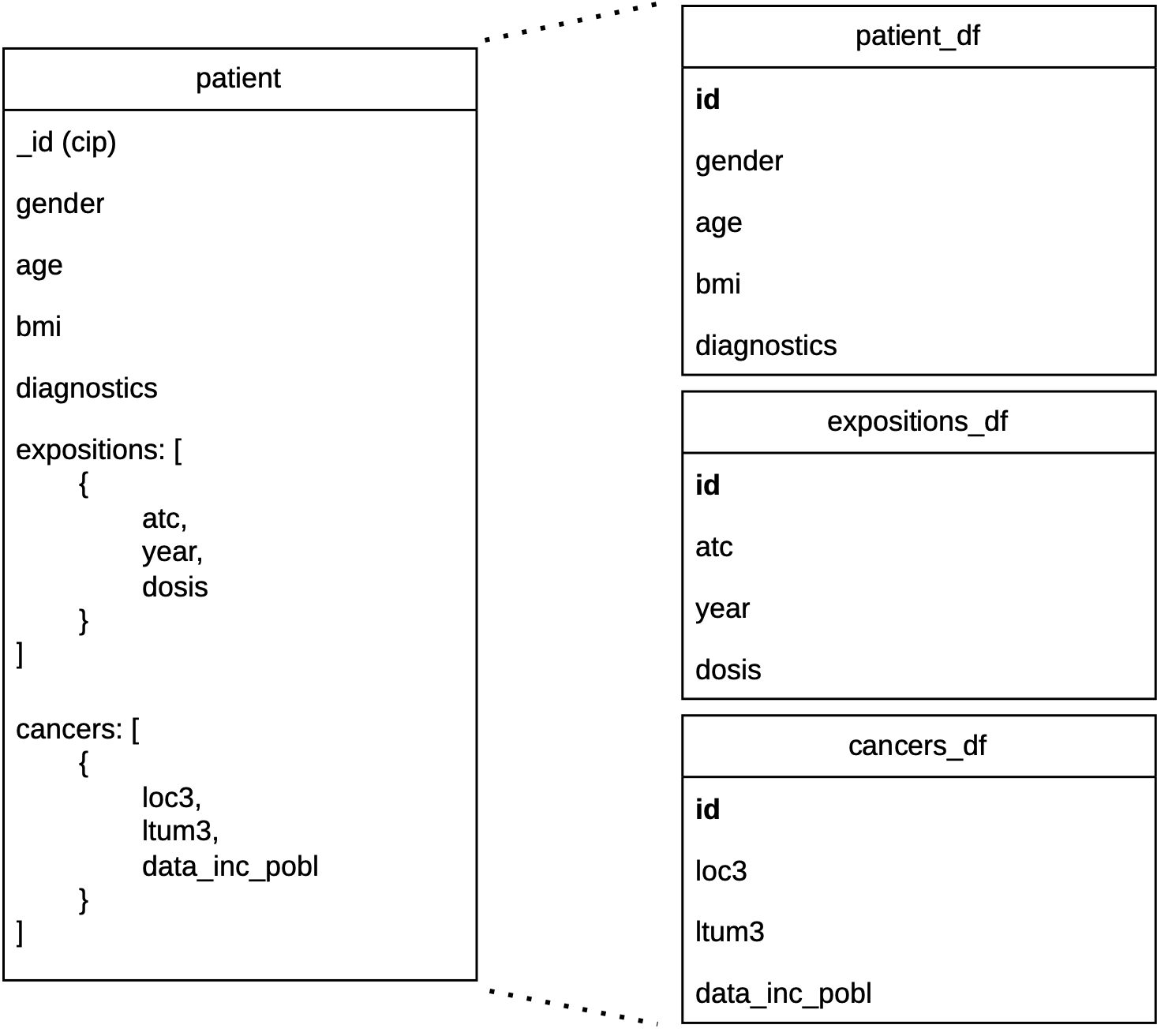
Solution
- Solution: split into 3 DataFrames:
patients,expositions,cancers
Memory Optimization
Findings
- Most patient features are invariant across combinations (age, BMI, diabetes…)
- Sorting the data and downcasting types can significantly reduce memory space.
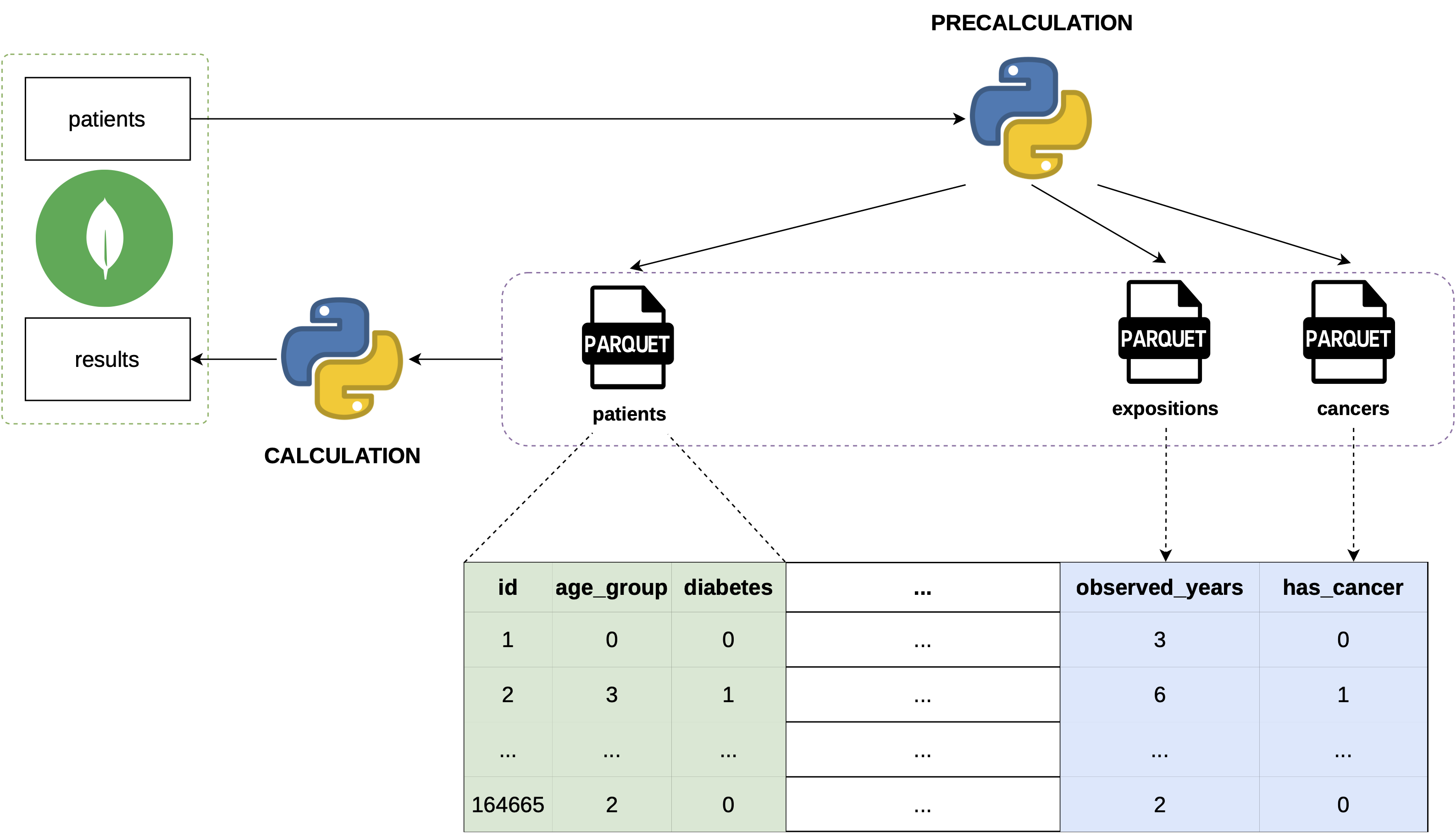
Proposal
- Precompute static features.
- Load data once into memory.
- Save shared data in Apache Parquet.
- Minimize queries to the database.
Upgrade
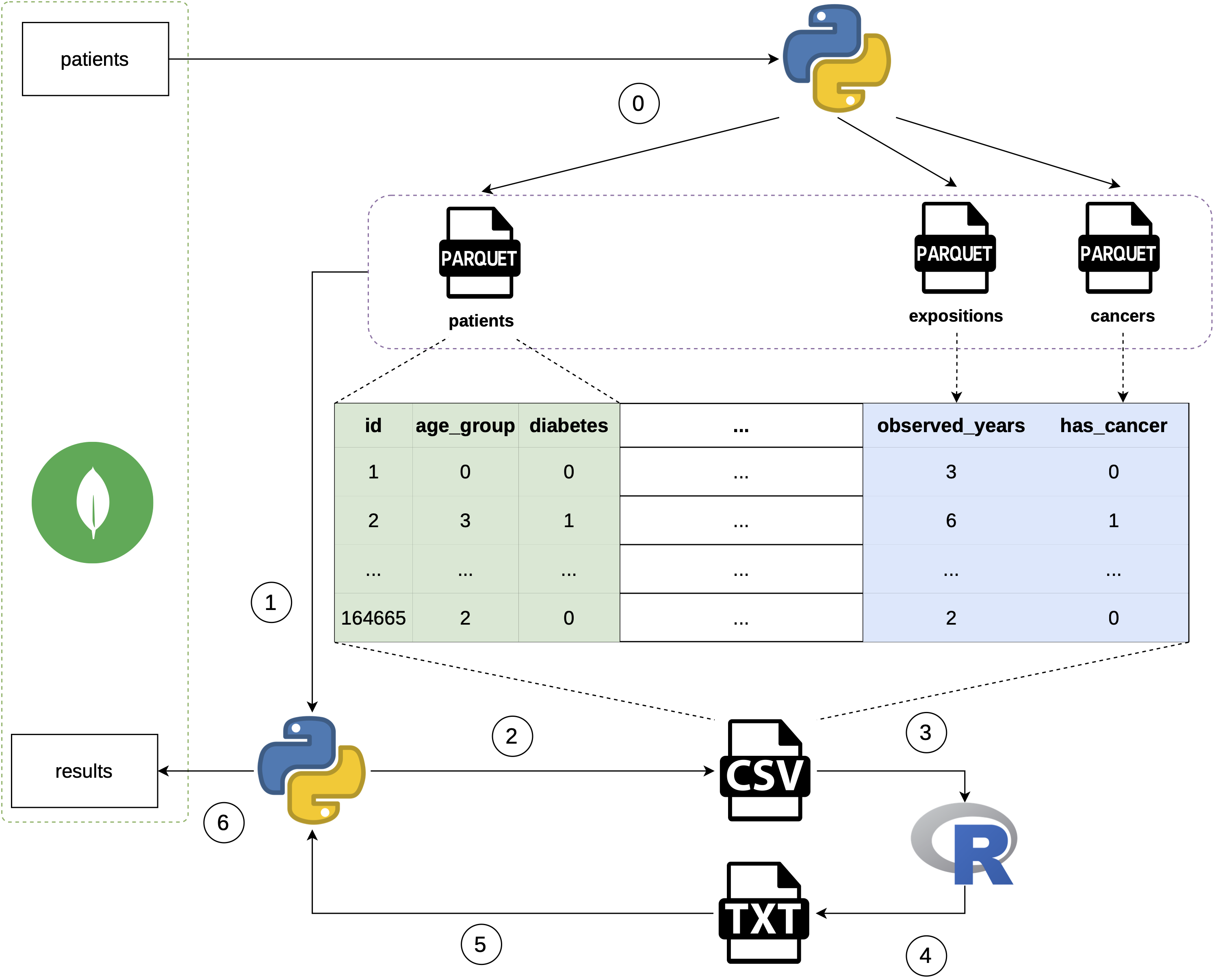
Mechanism
- Precalculate Dataframes
- Read the Dataframes
- CSV generation with features and the event column
- CSV reading and data preprocessing
- COXPH analysis (using a file as stdout)
- Read and parse the results
- Save structured results for later queries
Improvements
- Query-driven -> Reduce time to get data.
- Precalculate shared data -> Avoid repeated queries.
- Use Parquet files -> Efficient storage and fast access.
Code Optimizations1
Non-index-aware
Index-aware
Results
- Non-index-aware: 1.86 ms
- Index-aware: 247 μs
Using isin()
Using loc with prefill
Results
- Using
isin(): 3.25 ms - Using
locwith prefill: 464 μs
Summary of Optimizations
The total reduction in time is around 52 ms per combination. For 79,931 combinations, this results in a total time of ~1 hours.
Eliminating Communications
Problem
Disk I/O for inter-process communication (IPC) with R is a significant bottleneck.
Results
We reduce the processing time from 66 seconds to less than 1 second per combination.
| Function | Time (ms) |
|---|---|
get_cox_df |
52 |
calculate_cox_analysis |
776 |
parse_cox_analysis |
22 |
save_results |
21 |
Multithreading
Technical Insights
- Processes outperform threads for CPU-bound tasks -> Python’s Global Interpreter Lock (GIL) limits threads’ performance.
- Memory usage is higher with processes, which can lead to out-of-memory errors if too many processes are spawned.
- Threads are more efficient for I/O-bound tasks, allowing for faster startup and lower memory usage.
Hybrid Strategy
Since threads and processes are not mutually exclusive, we adopted a hybrid approach:
- Threads: Efficient for I/O and lightweight parallelism. Used with 2× CPU cores.
- Processes: Bypass GIL for CPU-bound tasks. Limited by available RAM.
Resource Calibration
The hybrid approach allows fine-tuned calibration of threads and processes, adapting to the device’s CPU and memory capacity. This ensures optimal throughput without exceeding hardware limits.
Task distribution
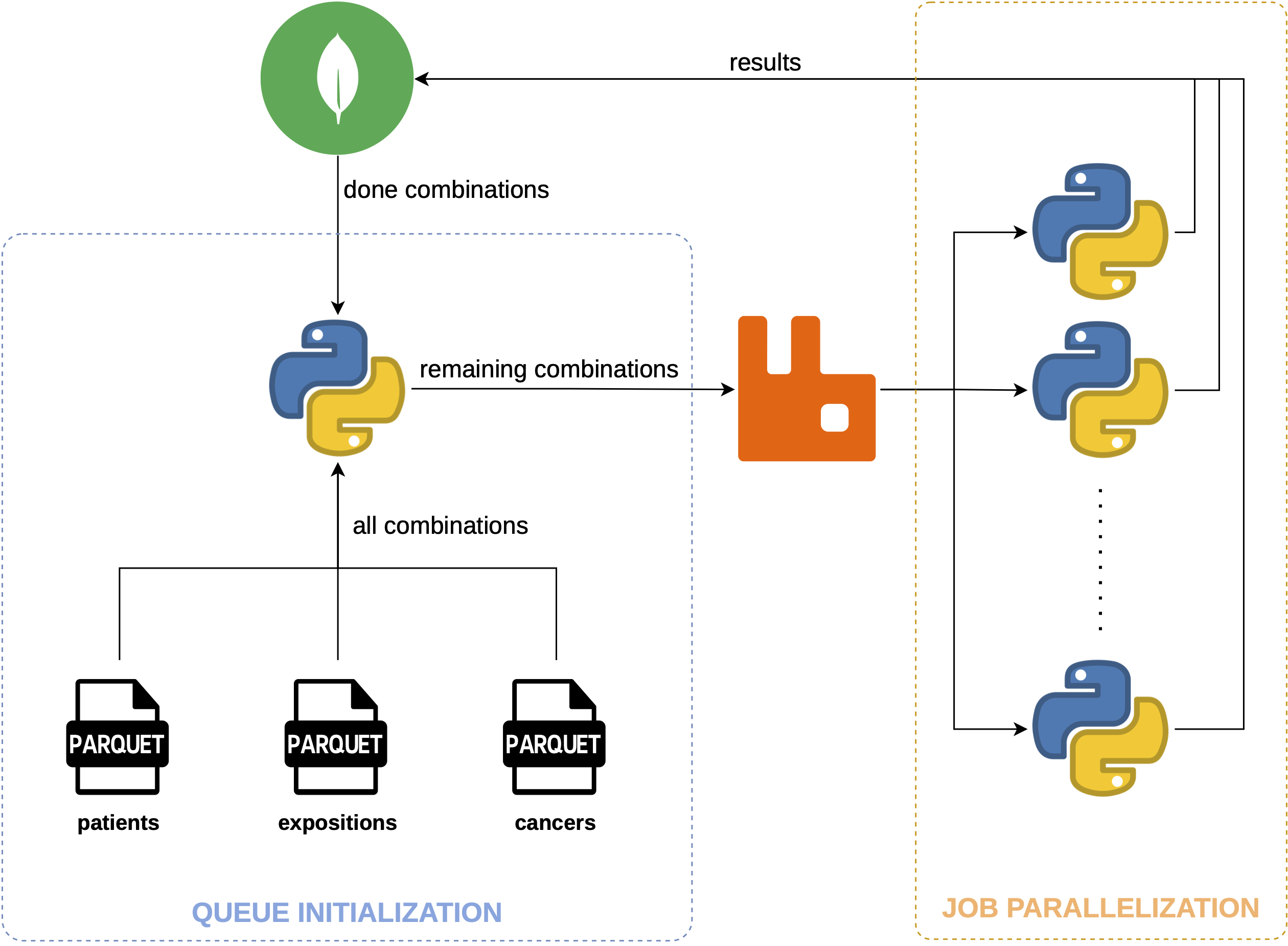
Architecture
- Task independence: Each task is a particular combination of medication and cancer type -> can be processed independently.
- Task Queue: Distributes tasks to worker processes (rabbitmq).
- Worker Processes: Can be configured to run on different machines, allowing for distributed computing.
- Task Management: Each worker fetches tasks from the queue, processes them, and returns results to the main process.
Deployment
Requirements
- Independent combinations to process.
- Scalable and reproducible execution.
Rationale1
| Feature | Traditional MPI Cluster | Kubernetes |
|---|---|---|
| Resource Allocation | Static (fixed per job) | Dynamic (per-task) |
| Scaling | Manual intervention required | Auto-scaling (HPA + Cluster) |
| Fault Tolerance | Job fails if worker crashes | Self-healing |
Scalability
Comparative Analysis
| Cloud | Instance type | Coremark | Workers | vCPUs | Tasks/s | Total time |
|---|---|---|---|---|---|---|
| GKE | e2-highcpu-4 | 51937 | 1 | 4 | 1.0 | 22h 12min |
| 2 | 8 | 1.9 | 11h 41min | |||
| 4 | 16 | 3.6 | 06h 10min | |||
| 8 | 32 | 7.0 | 03h 10min | |||
| c2d-highcpu-4 | 86953 | 4 | 16 | 17.0 | 01h 18min | |
| On-premise | opteron_6247 | 9634 | 1 | 10 | 0.4 | 2d 7h 30min |
| 2 | 20 | 0.88 | 1d 1h 13min | |||
| 4 | 40 | 2 | 11h 6min |
Conclusions
Key Optimizations
Schema Optimization
Query-driven design and better deserialization.Precomputation & Storage
Eliminated redundant calculations and migrated from CSV to Parquet for columnar efficiency.Compute Efficiency and Communication Overhead
Index-aware queries and optimized pipelines.Parallel Execution
Hybrid threading/multiprocessing to maximize resource utilization.Distributed Scaling
Kubernetes-orchestrated workers with queue-based load balancing.
Take Home Messages
- 61 Days → ~Hours
Computational throughput improved through systematic optimization.
References

CMMSE 2025